Software Exercise 1.1: Using a Pre-compiled Binary¶
In this exercise, we will run a job using a downloaded, pre-compiled binary. This exercise should take 10-15 minutes.
Background¶
This is the simplest scenario for using a particular software program on the Open Science Grid - downloading a pre-compiled binary and using it to run jobs.
Our Software Example¶
The software we will be using for this example is a common tool for aligning genome and protein sequences against a reference database, the BLAST program.
-
Search the internet for the BLAST software. Searches might include "blast executable or "download blast software". Hopefully these searches will lead you to a BLAST website page that looks like this:
-
Click on the title that says "Download BLAST" and then look for the link that has the latest installation and source code. You should end up on a page with a list of each version of BLAST that is available for different operating systems.
-
We could download the source and compile it ourselves, but instead, we're going to use one of the pre-built binaries. Before proceeding, look at the list of downloads and try to determine which one you want.
-
Based on our operating system, we want to use the Linux binary, which is labelled with the
x64-linuxsuffix.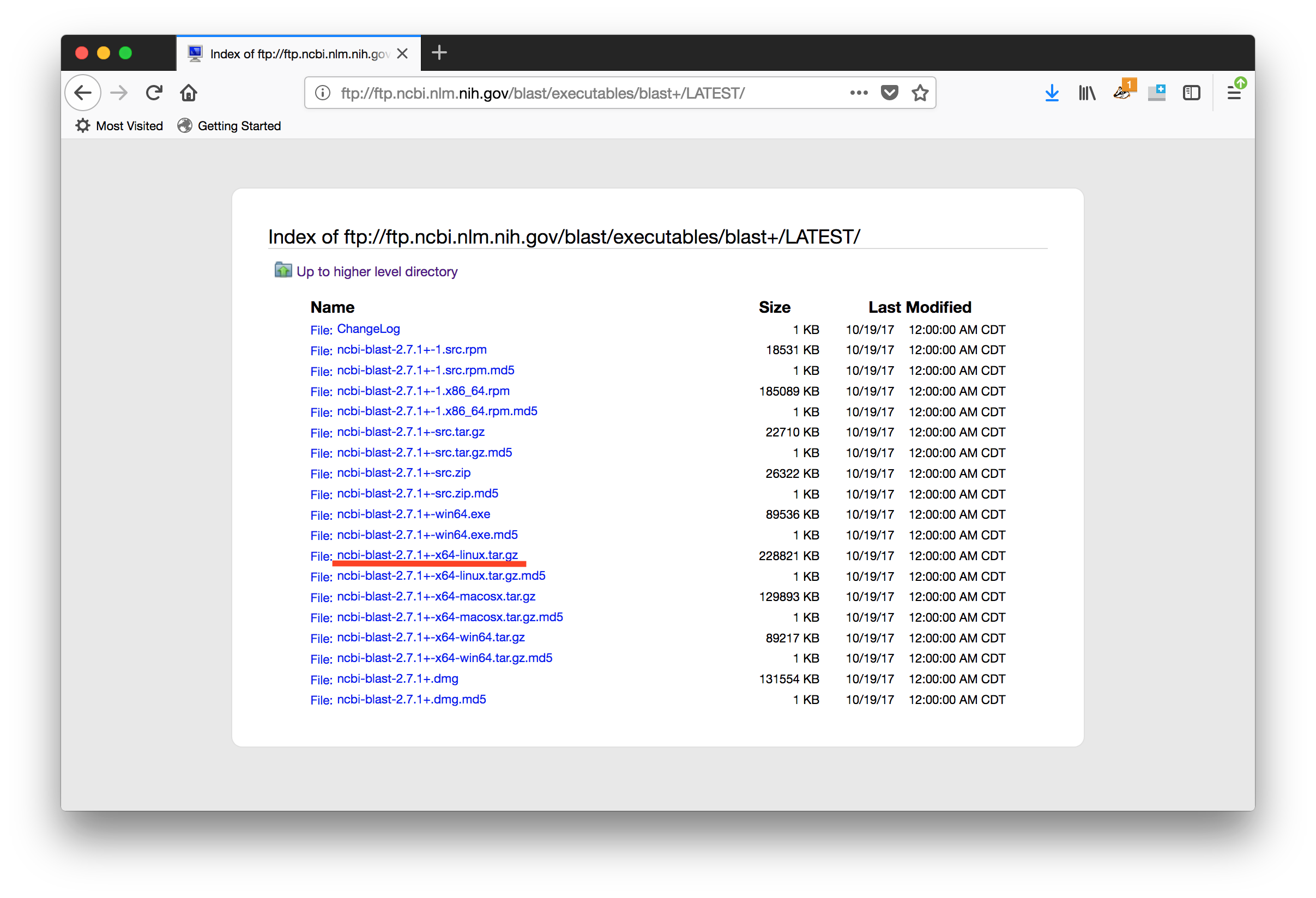
All the other links are either for source code or other operating systems.
-
While logged into
login04.osgconnect.net, create a directory for this exercise. Then download the appropriatetar.gzfile and un-tar/decompress it it. If you want to do this all from the command line, the sequence will look like this (usingwgetas the download command.)user@login $ wget https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ncbi-blast-2.12.0+-x64-linux.tar.gz user@login $ tar -xzf ncbi-blast-2.12.0+-x64-linux.tar.gz -
We're going to be using the
blastxbinary in our job. Where is it in the directory you just decompressed?
Copy the Input Files¶
To run BLAST, we need an input file and reference database. For this example, we'll use the "pdbaa" database, which contains sequences for the protein structure from the Protein Data Bank. For our input file, we'll use an abbreviated fasta file with mouse genome information.
-
Download these files to your current directory:
username@login $ wget http://proxy.chtc.wisc.edu/SQUID/osgschool21/pdbaa.tar.gz username@login $ wget http://proxy.chtc.wisc.edu/SQUID/osgschool21/mouse.fa -
Untar the
pdbaadatabase:username@login $ tar -xzf pdbaa.tar.gz
Submitting the Job¶
We now have our program (the pre-compiled blastx binary) and our input
files, so all that remains is to create the submit file. The form of a
typical blastx command looks something like this:
blastx -db <database_dir/prefix> -query <input_file> -out <output_file>
-
Copy a submit file from one of the Day 2 exercises to use for this exercise.
-
Think about which lines you will need to change or add to your submit file in order to submit the job successfully. In particular:
- What is the executable?
- How can you indicate the entire command line sequence above?
- Which files need to be transferred in addition to the executable?
- Does this job require a certain type of operating system?
- Do you have any idea how much memory or disk to request?
-
Try to answer these questions and modify your submit file appropriately.
-
Once you have done all you can, check your submit file against the lines below, which contain the exact components to run this particular job.
-
The executable is
blastx, which is located in thebindirectory of our downloaded BLAST directory. We need to use theargumentsline in the submit file to express the rest of the command.executable = ncbi-blast-2.12.0+/bin/blastx arguments = -db pdbaa/pdbaa -query mouse.fa -out results.txt -
The BLAST program requires our input file and database, so they must be transferred with
transfer_input_files.transfer_input_files = pdbaa, mouse.fa -
Let's assume that we've run this program before, and we know that 1GB of disk and 1GB of memory will be MORE than enough (the 'log' file will tell us how accurate we are, after the job runs):
request_memory = 1GB request_disk = 1GB -
Because we downloaded a Linux-specific binary, we need to request machines that are running Linux.
requirements = (OSGVO_OS_STRING == "RHEL 7")
-
-
Submit the blast job using
condor_submit. Once the job starts, it should run in just a few minutes and produce a file calledresults.txt.